Bio
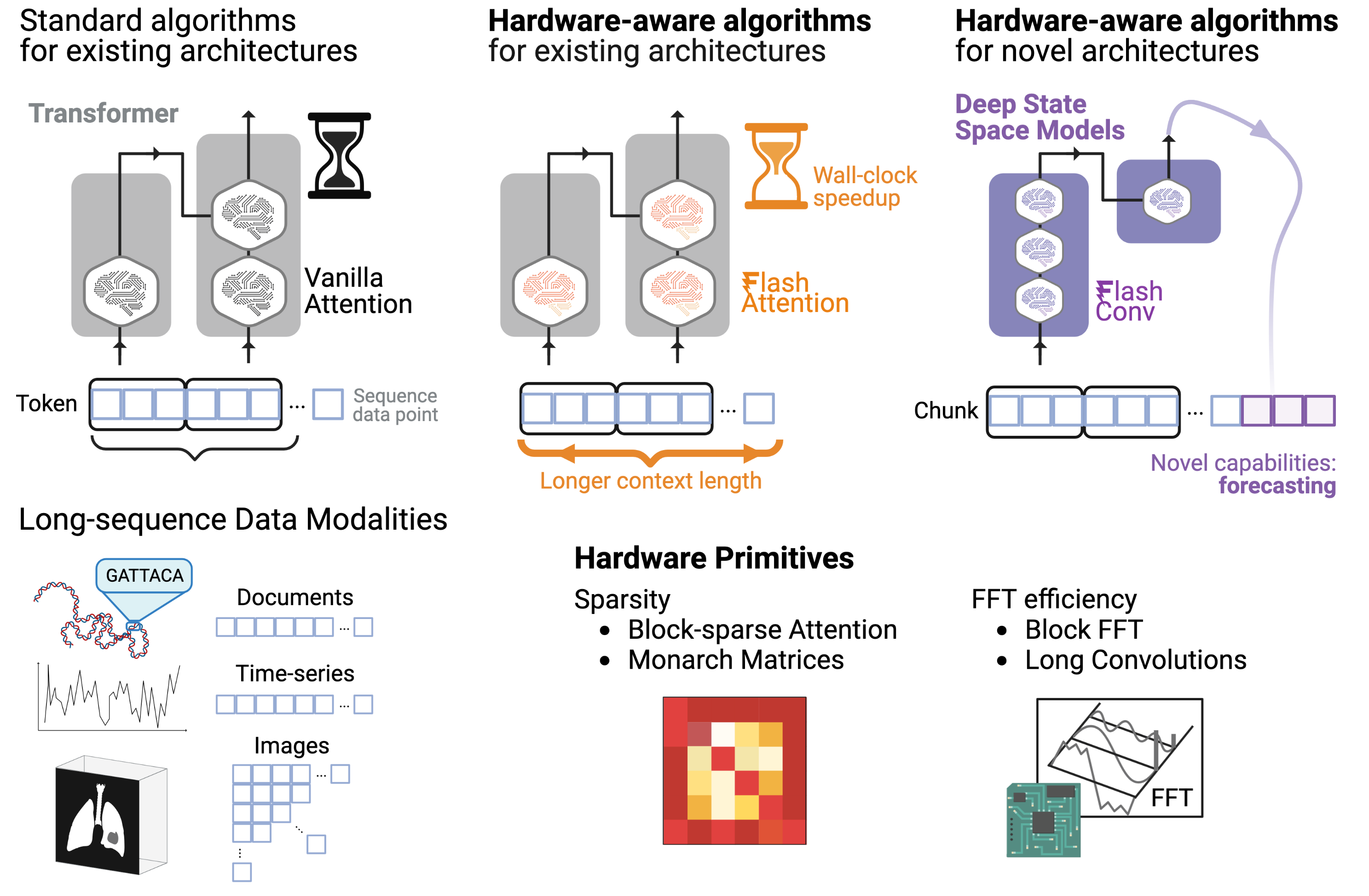
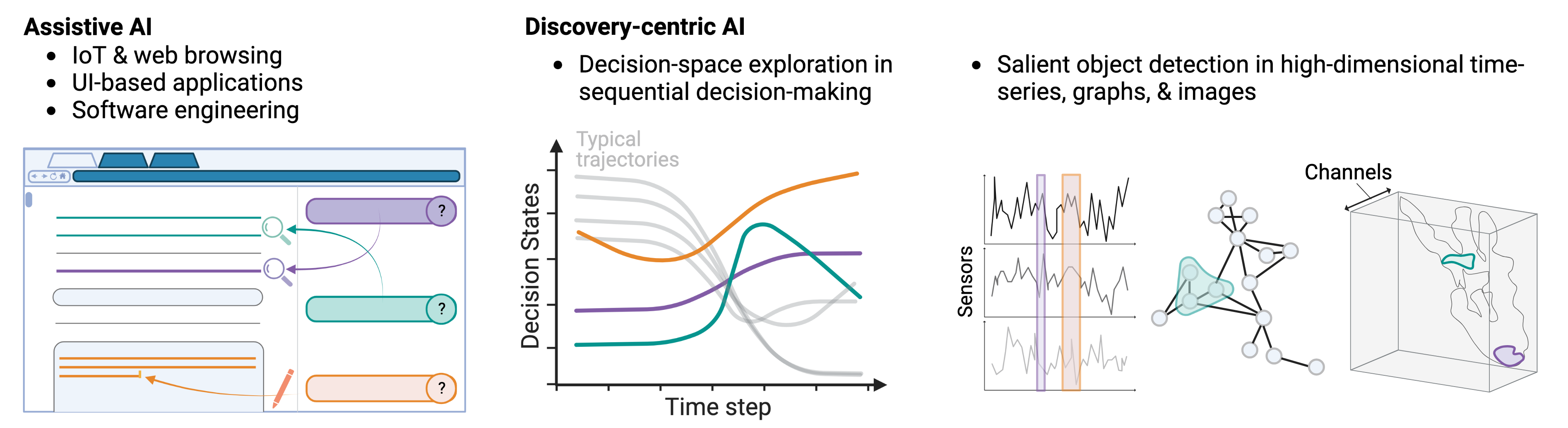
I am a Member of the Technical Staff at Radical Numerics, an SF- & Tokyo-based startup building frontier biological sequence models. My research efforts focus on (a) building modeling primitives for long-context attribution and interpretability, (b) test-time model steering, and (c) methods applications for AI-driven knowledge acquisition — ultimately aiding in our understand of biological systems and the natural world.
I completed my PhD in Biomedical Informatics at the Stanford's department of Biomedical Data Science and Stanford AI Lab (SAIL), where I was extremely fortunate to be co-advised and mentored by Parag Mallick (Radiology) and Christopher Ré (CS). My undergraduate training was in Applied Mathematics and Bioengineering at University of California, Berkeley. My CV can be found here.
My name is most similarly pronounced as Batman's city of residence, "Gotham" (i.e. GAW-thum). There are many intonation variants for my Sanskrit-originating name across India, but this pronunciation is the closest to the North Indian variant. I also go by "Machi" if that's easier.
Research Artifacts
Before working on biological sequence models, I've had the pleasure of building models for multiple token types and modalities in the biomedical domain:
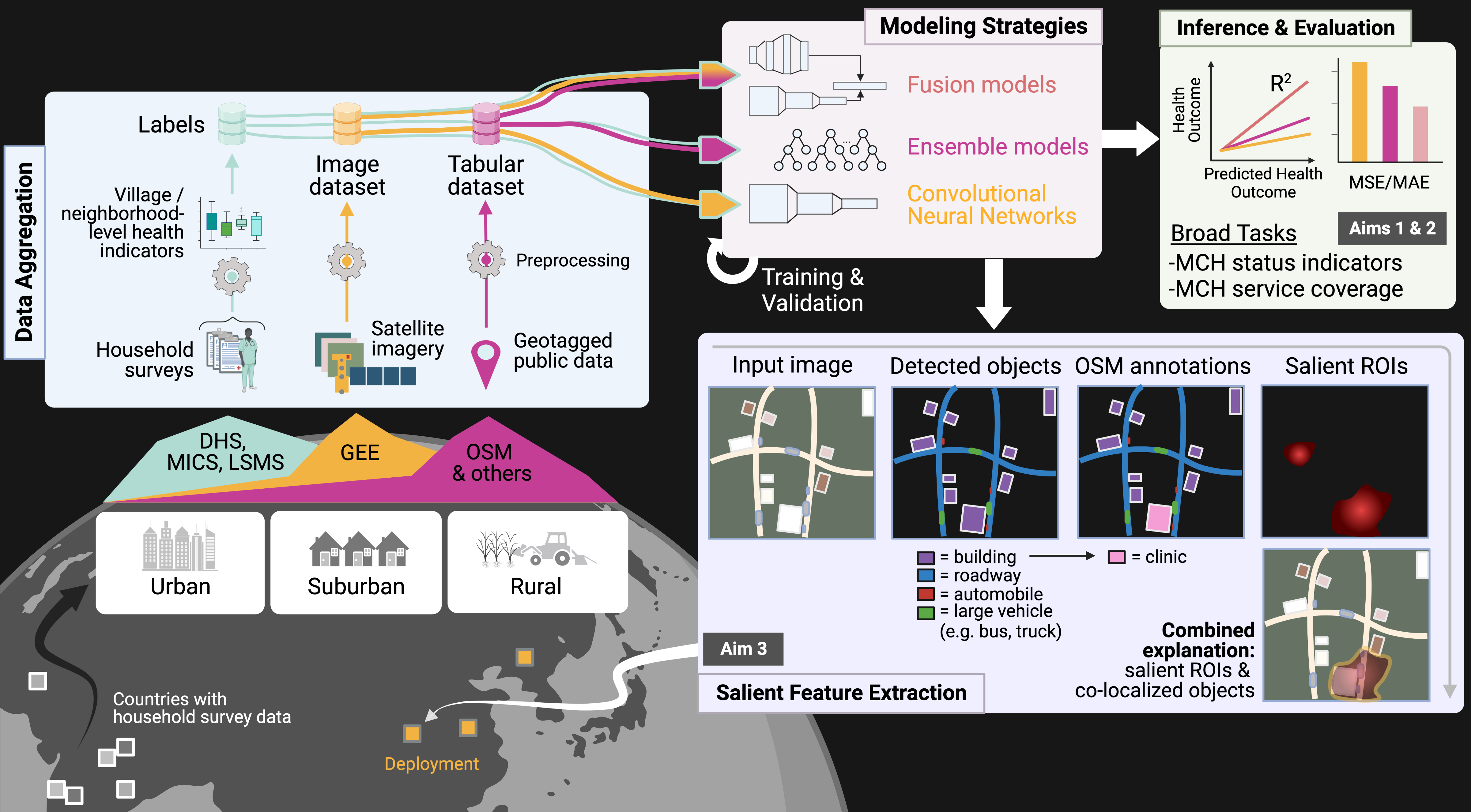
- pixels in images — pathology slides, edge devices, video frames, satellite imagery
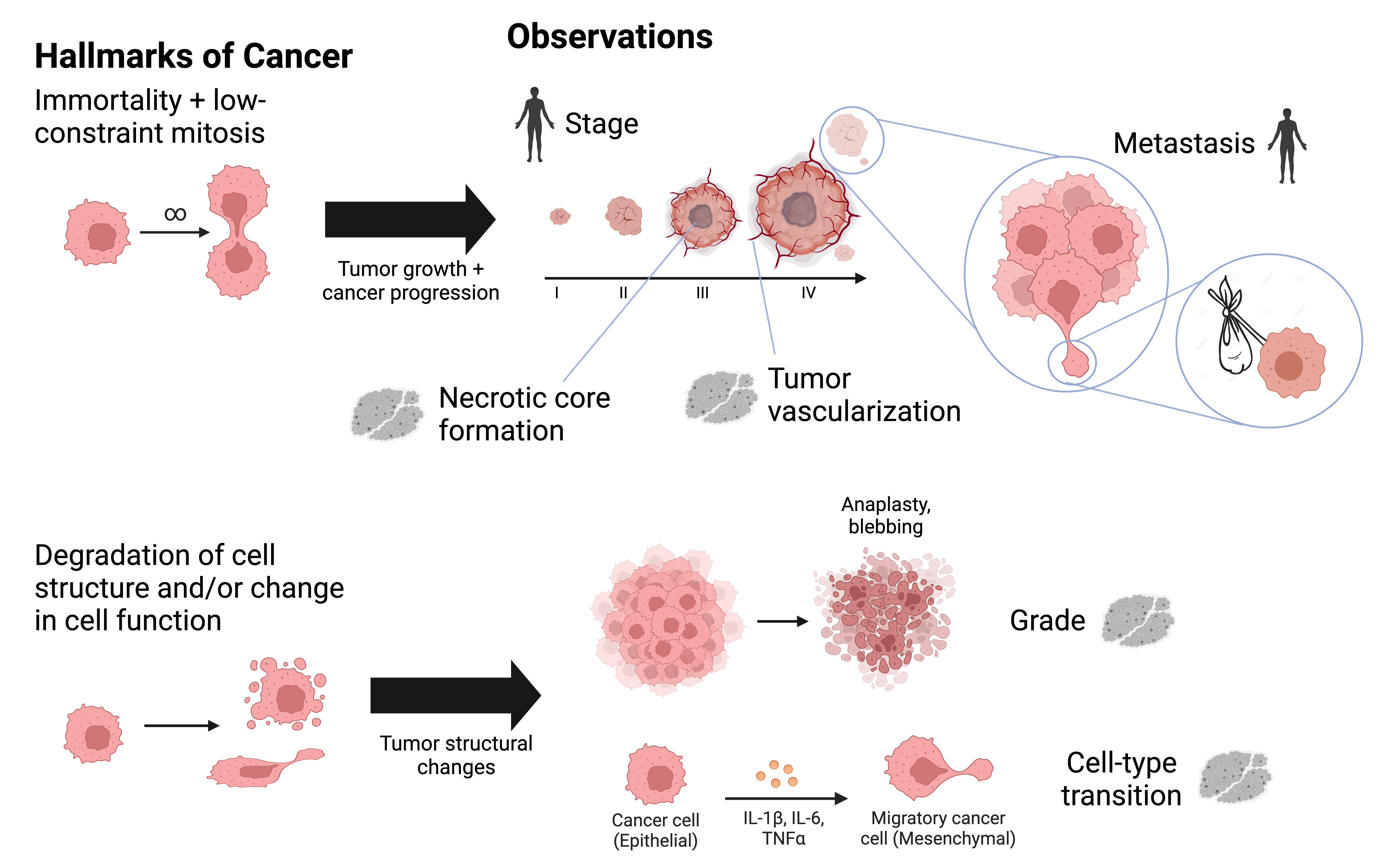
- cells in tumors — spatial proteomics
- text spans in natural language — scientific literature, EHRs
- monomers in biological sequences — DNA, proteins
Smarter by Design: How SmarterDx Engineered Clinical AI for the Complexity of Healthcare
Company whitepaper (2025)
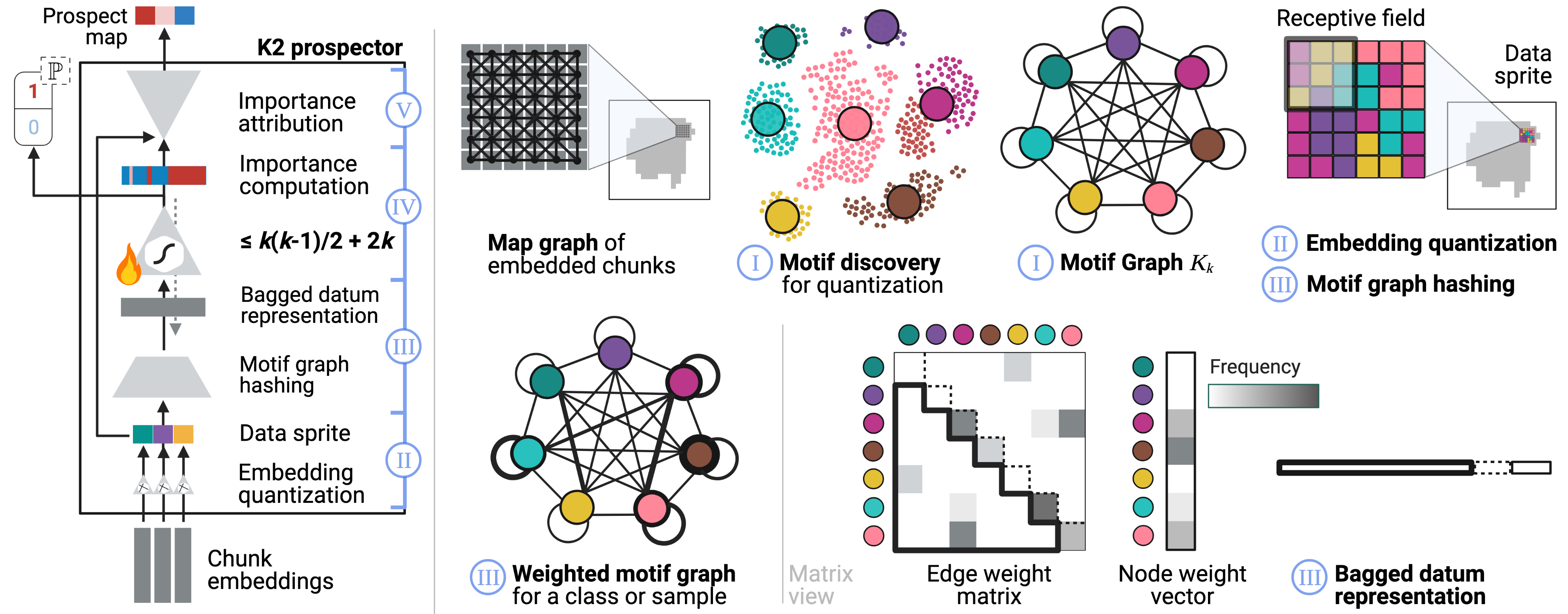
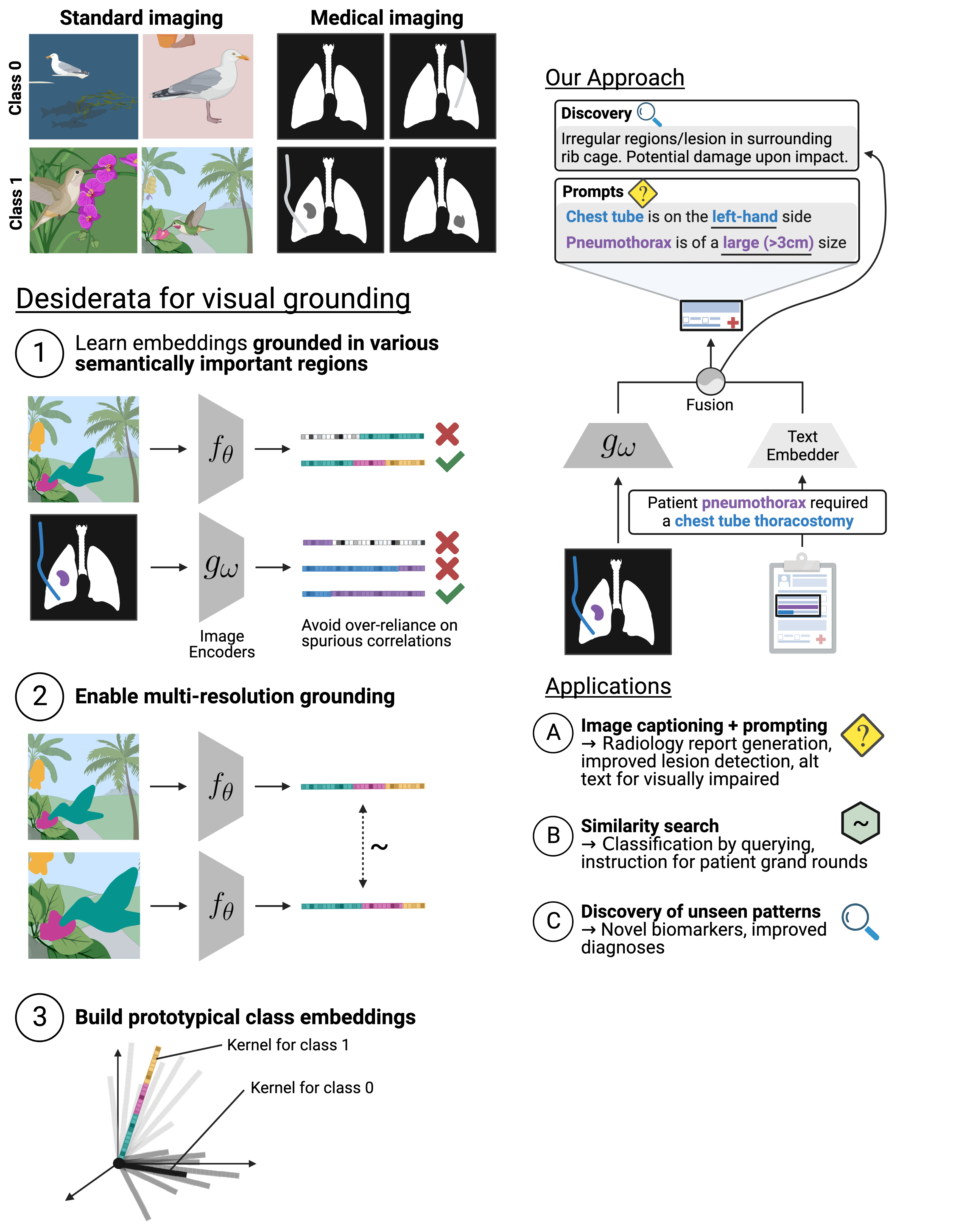
Prospector Heads: Generalized Feature Attribution for Large Models & Data
Gautam Machiraju, Alexander Derry, Arjun Desai, Neel Guha, Amir-Hossein Karimi, James Zou, Russ Altman, Christopher Ré, Parag Mallick
International Conference on Machine Learning (ICML), 2024
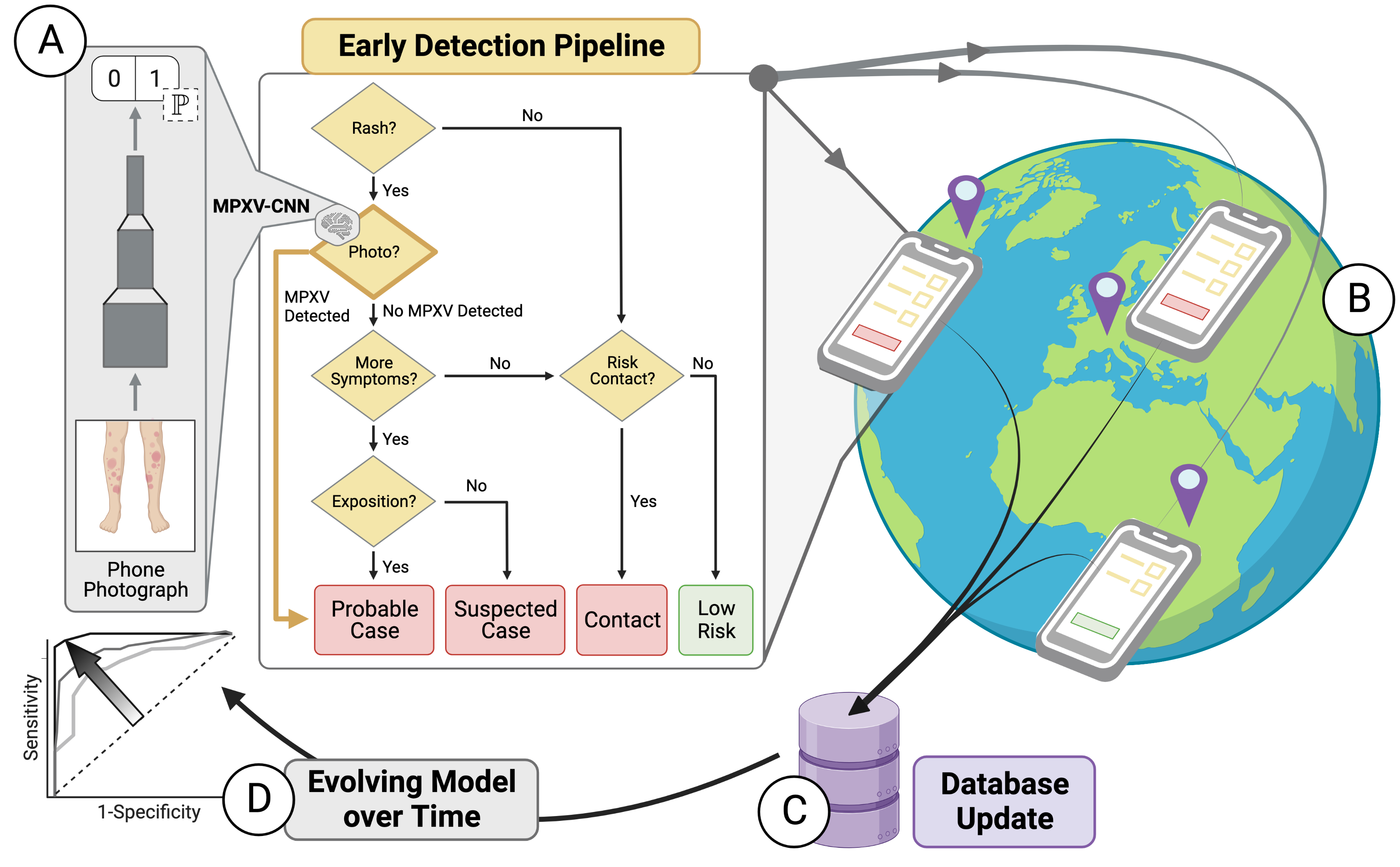
Development and Evaluation of an Image-based Deep Learning Algorithm to Classify Skin Lesions from Mpox Virus Infection
Alexander Henry Thieme, Yuanning Zheng, Gautam Machiraju, et al.
Nature Medicine (2023)
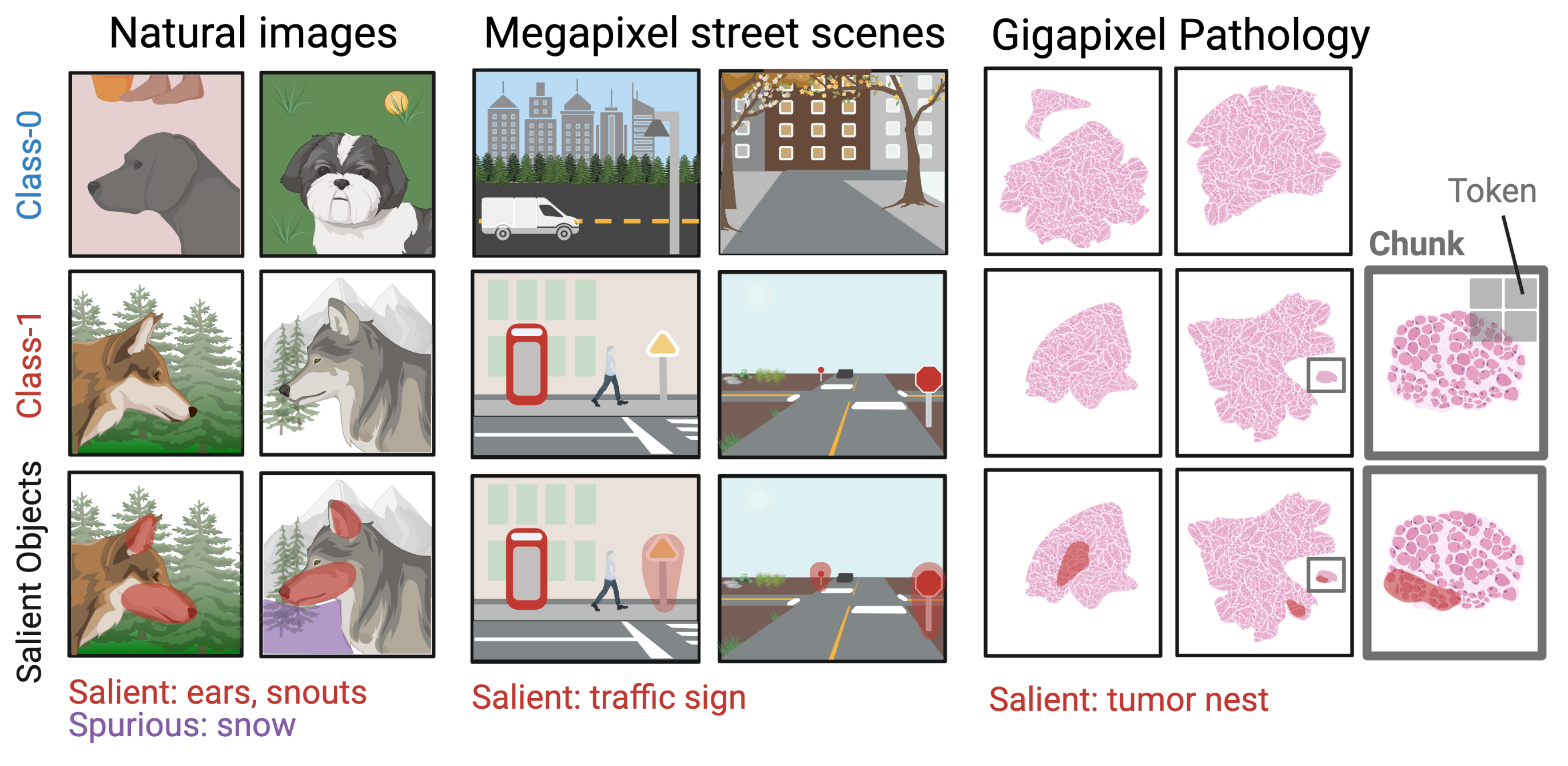
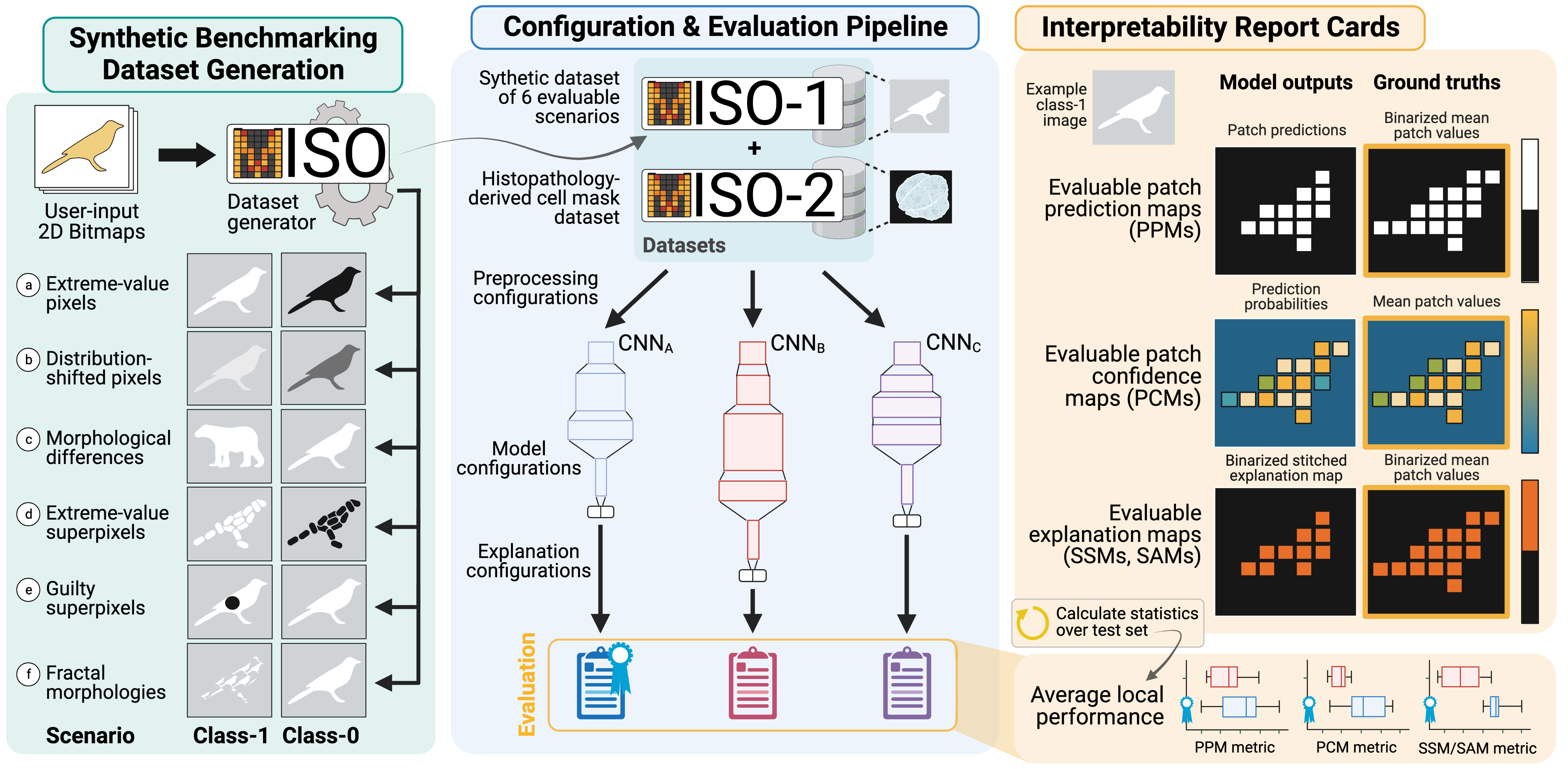
A Dataset Generation Framework for Evaluating Megapixel Image Classifiers & their Explanations
Gautam Machiraju, Sylvia Plevritis, Parag Mallick
European Conference on Computer Vision (ECCV), 2022
Developing Machine Learning Models to Personalize Care Levels among Emergency Room Patients for Hospital Admission
Minh Nguyen, Conor Corbin, Tiffany Eulalio, Nicolai Ostberg, Gautam Machiraju, Ben Marafino, Michael Baiocchi, Christian Rose, Jonathan Chen
Journal of the American Medical Informatics Association (2021)
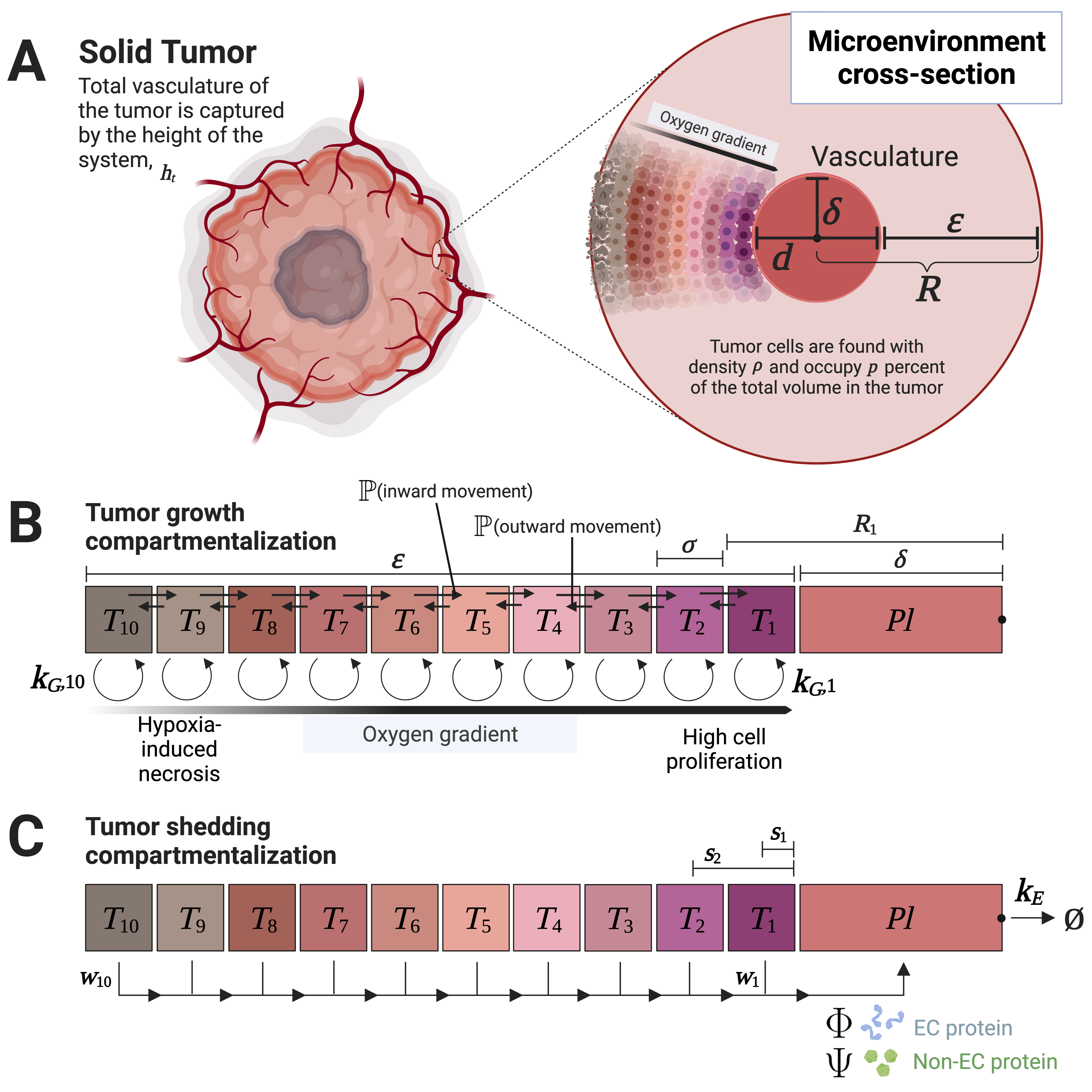
Multicompartment Modeling of Protein Shedding Kinetics During Vascularized Tumor Growth
Gautam Machiraju, Parag Mallick, Hermann Frieboes
Nature Scientific Reports (2020)
Etcetera
In my creative down-time, I enjoy exploring graphic/visual design, architectural photography, and sound design.
Acknowledgement
This website uses the website design and template by Martin Saveski. Some stylisitc alterations were made with inspiration from Tatsunori Hashimoto.