Bio
I am an ML Research Scientist at SmarterDx, an NYC-based healthAI startup working to better capture differential diagnoses from EHRs. My research efforts focus on long-context retrieval and LLM interpretability and steering.
I recently completed my PhD in Biomedical Informatics at the Stanford AI Lab (SAIL) and the department of Biomedical Data Science. My previous and current work centers around building data copilots — AI systems that augment our capabilities in (1) making rational and evidence-based decisions, (2) discovering and acquiring knowledge, and (3) generating new data given our design preferences. Our copilots were built to understand the natural world, particularly for biological and physical systems. I was extremely fortunate to be co-advised and mentored by Parag Mallick (Radiology) and Christopher Ré (CS).
My name is most similarly pronounced as Batman's city of residence, "Gotham" (i.e. GAW-thum). There are many intonation variants for my Sanskrit-originating name across India, but this pronunciation is the closest to the North Indian variant. I also go by "Machi" if that's easier.
Research
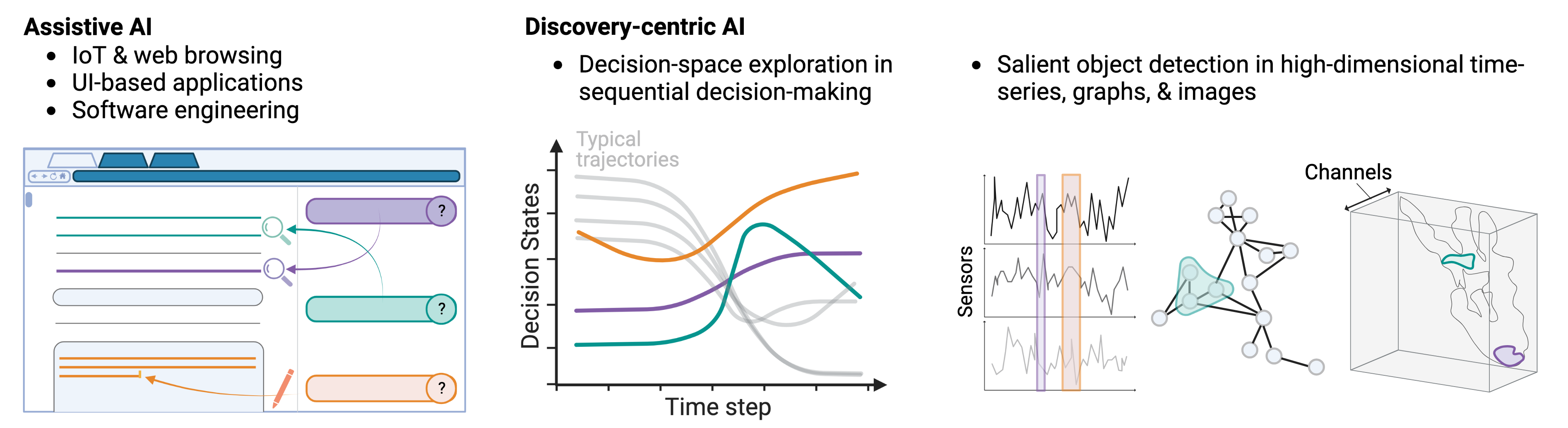
My research focuses on building data copilots, or AI systems that have the following interrelated capabilities:
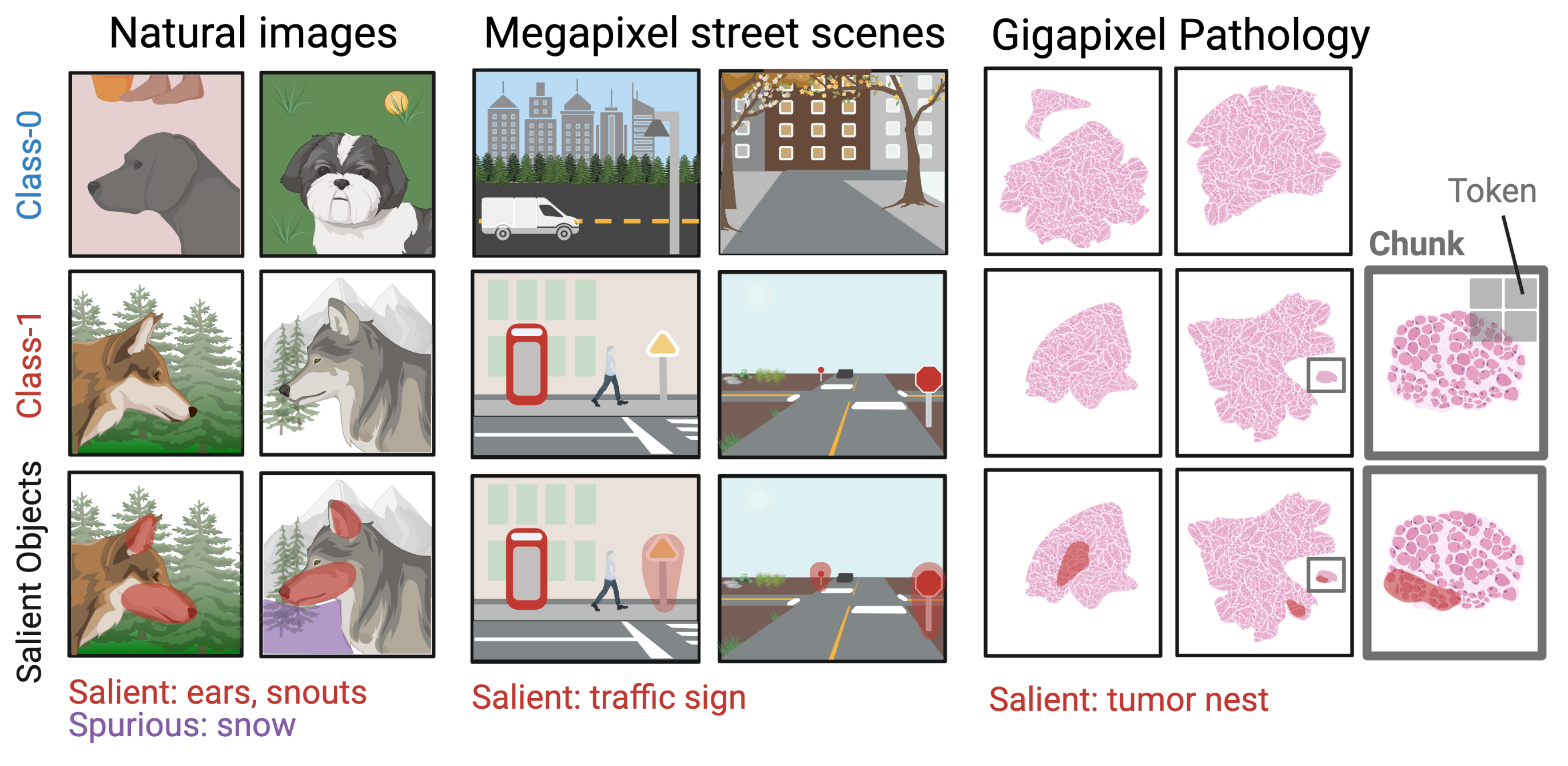
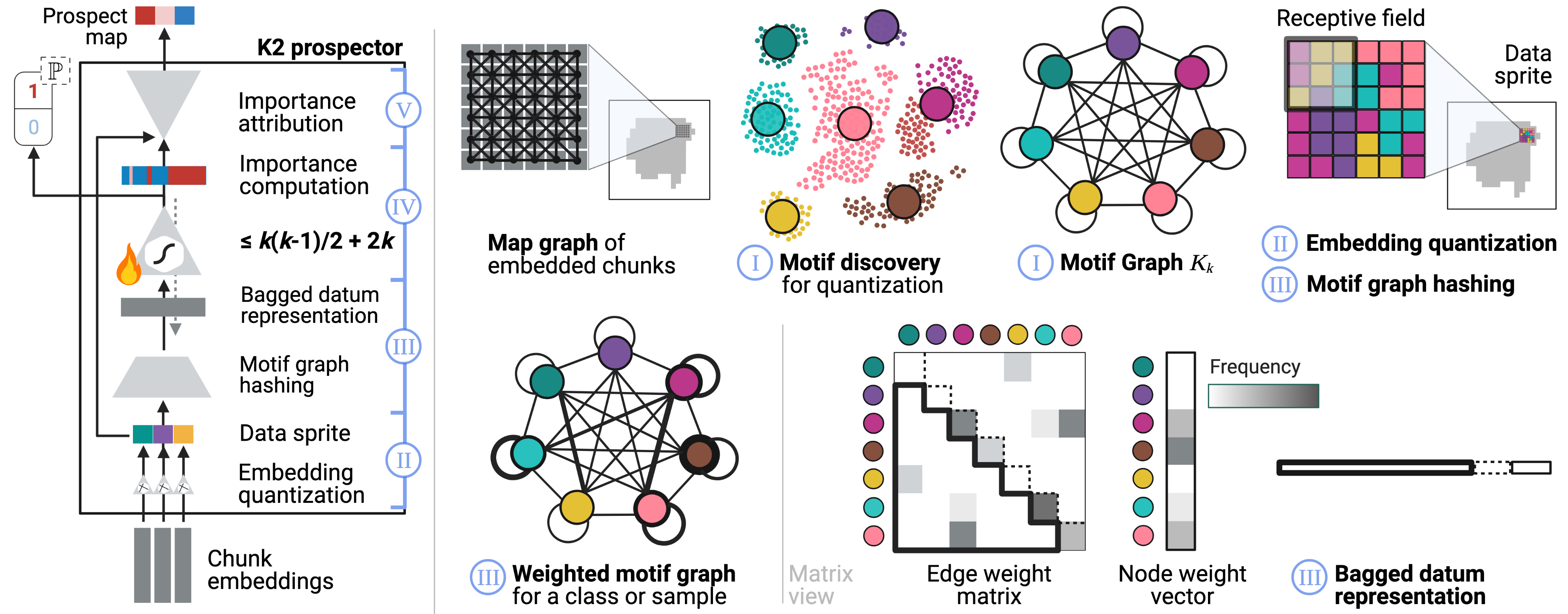
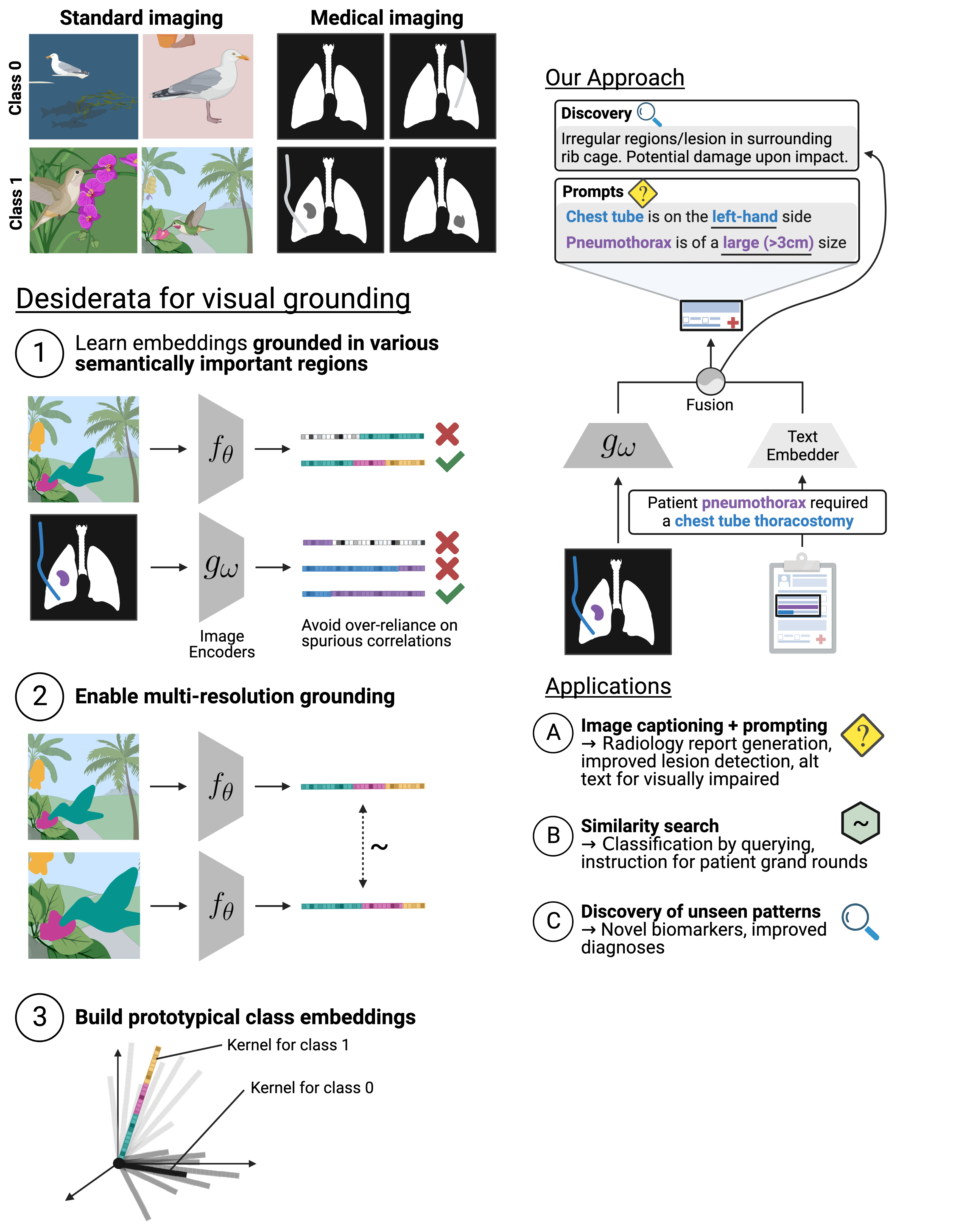
- Evidence-based & rational decision-making — pinpointing what is important (evidence) within inputs for a model's outputs, as well as why and how those outputs were created (rationale). Our work has built automated evidence-verification systems with synthetic data [ECCV'22] and new equippable, GPT-inspired architectural blocks that reveal AI evidence and rationale in text documents, pathology images, and protein sequences & structures [IMLH'23,ICML'24].
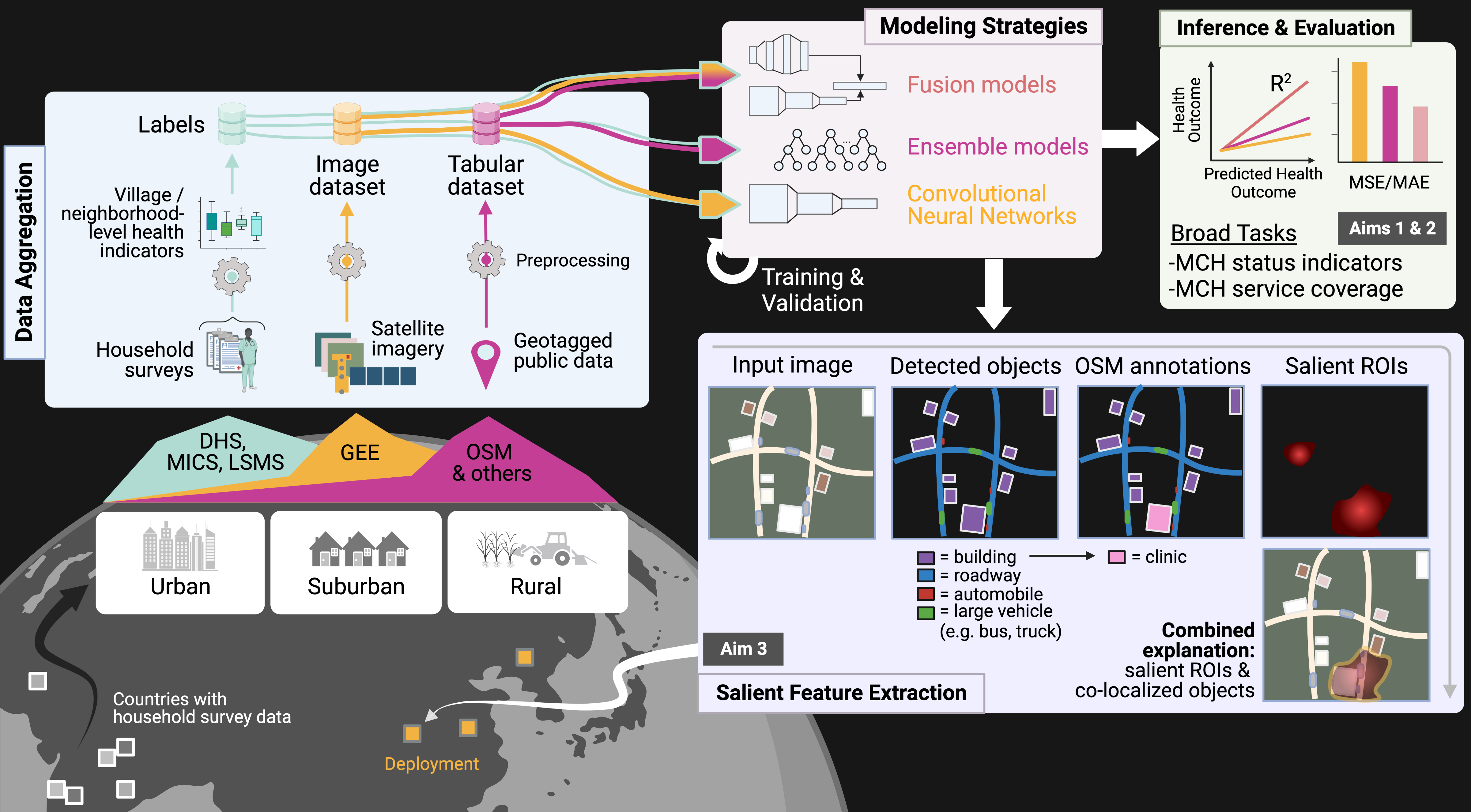
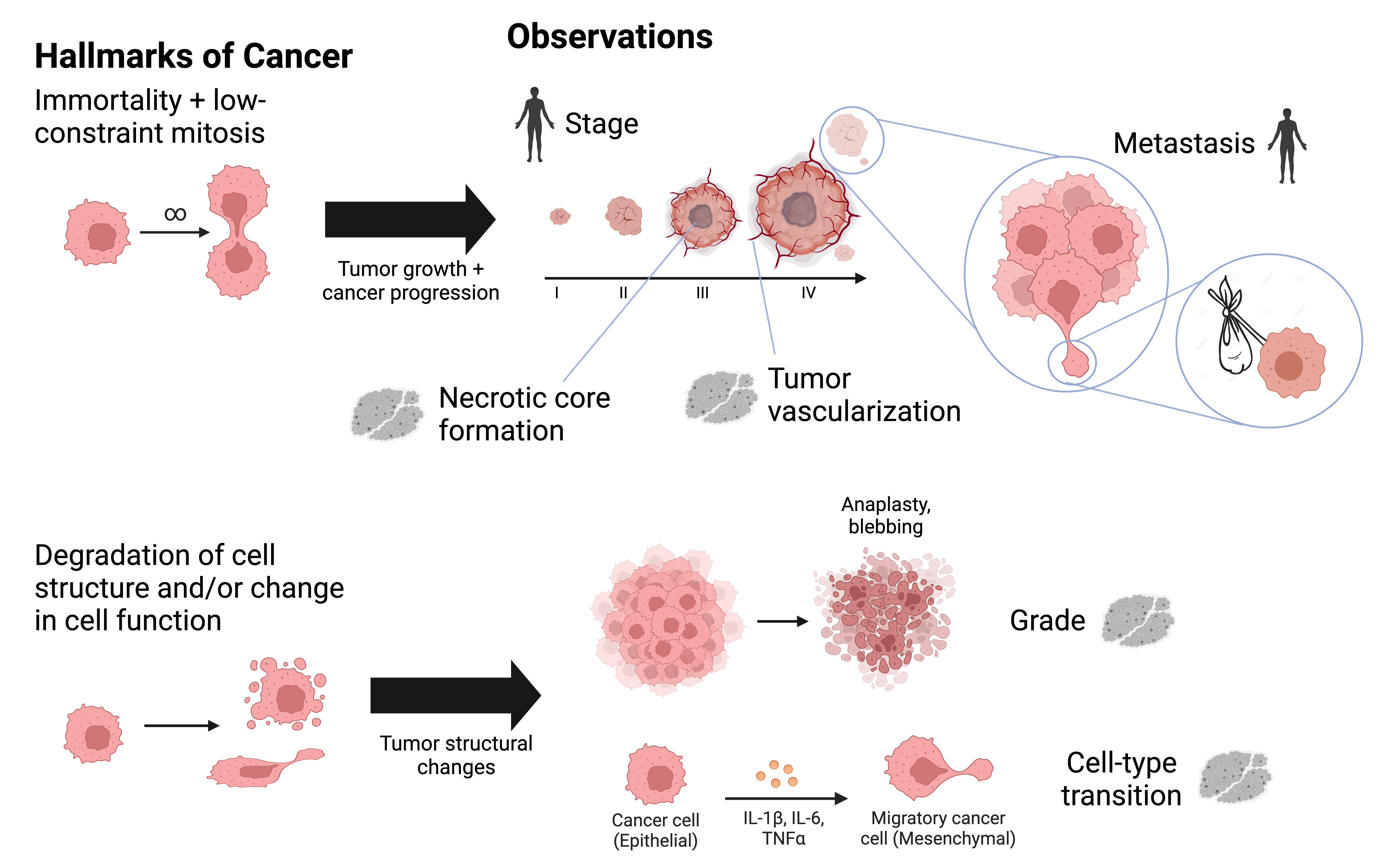
- Knowledge discovery & acquisition — identifying new information and ensuring it is human-comprehensible. We used model rationale from capability (1) in scientific deployments to discover tumor regions associated with aggressive cancers via spatial proteomics data and single-cell encoders [upcoming preprint]. We are also making deployments in global health and justice, adapting vision-language models to mine covariates from satellite imagery to combat human labor trafficking in the Amazon Rainforest. Finally, we are using protein language models (PLMs) to learn the "functional language" of synthetic and natural proteoforms.
- Controlled LLM generation — using weak labels and machine-extracted knowledge (e.g. memory banks of salient motifs) to guide LLMs toward desired data generation at test-time. Primary applications are rooted in protein sequence design [upcoming preprint].
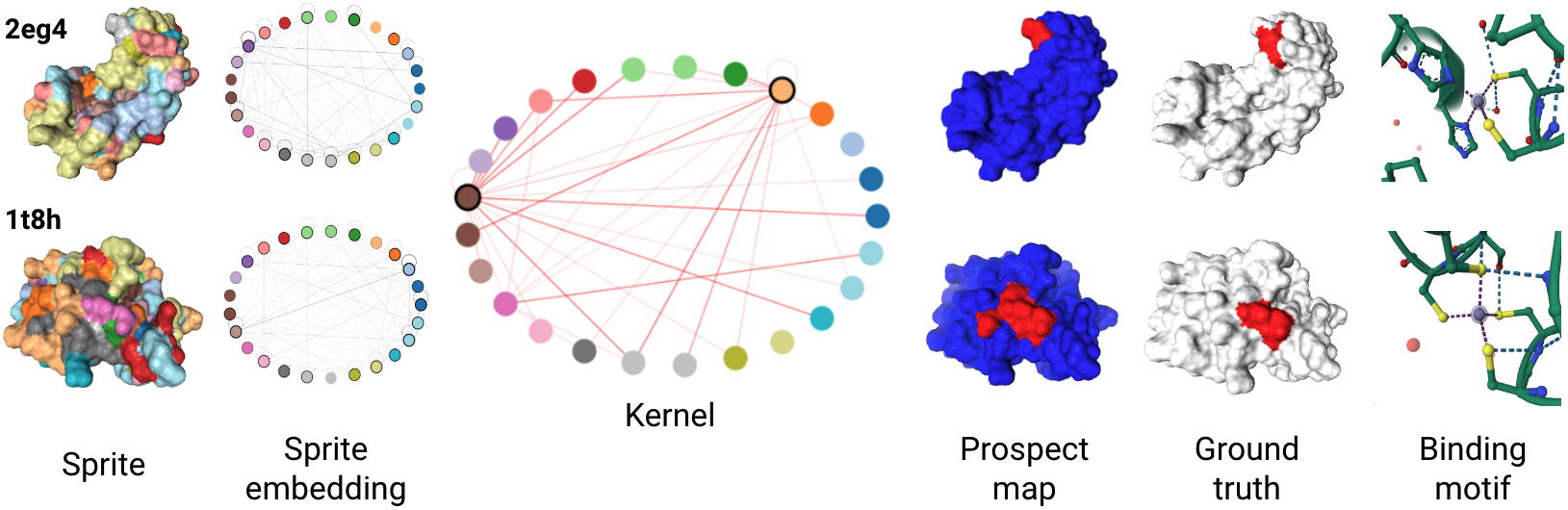
Projects & Artifacts
Stay tuned for future publications and writings on:
- Data copilots for spatial biology: in silico discovery of biomarkers of aggressive cancers
- A new form of controlled generation that incorporates weak supervision, motivated by protein design
- A review on spatial statistics for spatial biology
- A call to action for evidence-based and rational AI systems in biomedicine
- A lecture introducing modern ML to life scientists: the history, developments, and opportunities
Most recent publications on Google Scholar.
Grammar Matters: Grammatical Templates Improve Language Model Fine-Tuning for Biomedical Relation Extraction
Varun Tandon, Gautam Machiraju, Parag Mallick
In review.
Prospector Heads: Generalized Feature Attribution for Large Models & Data
Gautam Machiraju, Alexander Derry, Arjun Desai, Neel Guha, Amir-Hossein Karimi, James Zou, Russ Altman, Christopher Ré, Parag Mallick
International Conference on Machine Learning (ICML), 2024
Prospectors: Leveraging Short Contexts to Mine Salient Objects in High-dimensional Imagery
Gautam Machiraju, Arjun Desai, James Zou, Christopher Ré, Parag Mallick
International Conference on Machine Learning (ICML) 3rd workshop on Interpretable Machine Learning for Healthcare (IMLH) 2023.
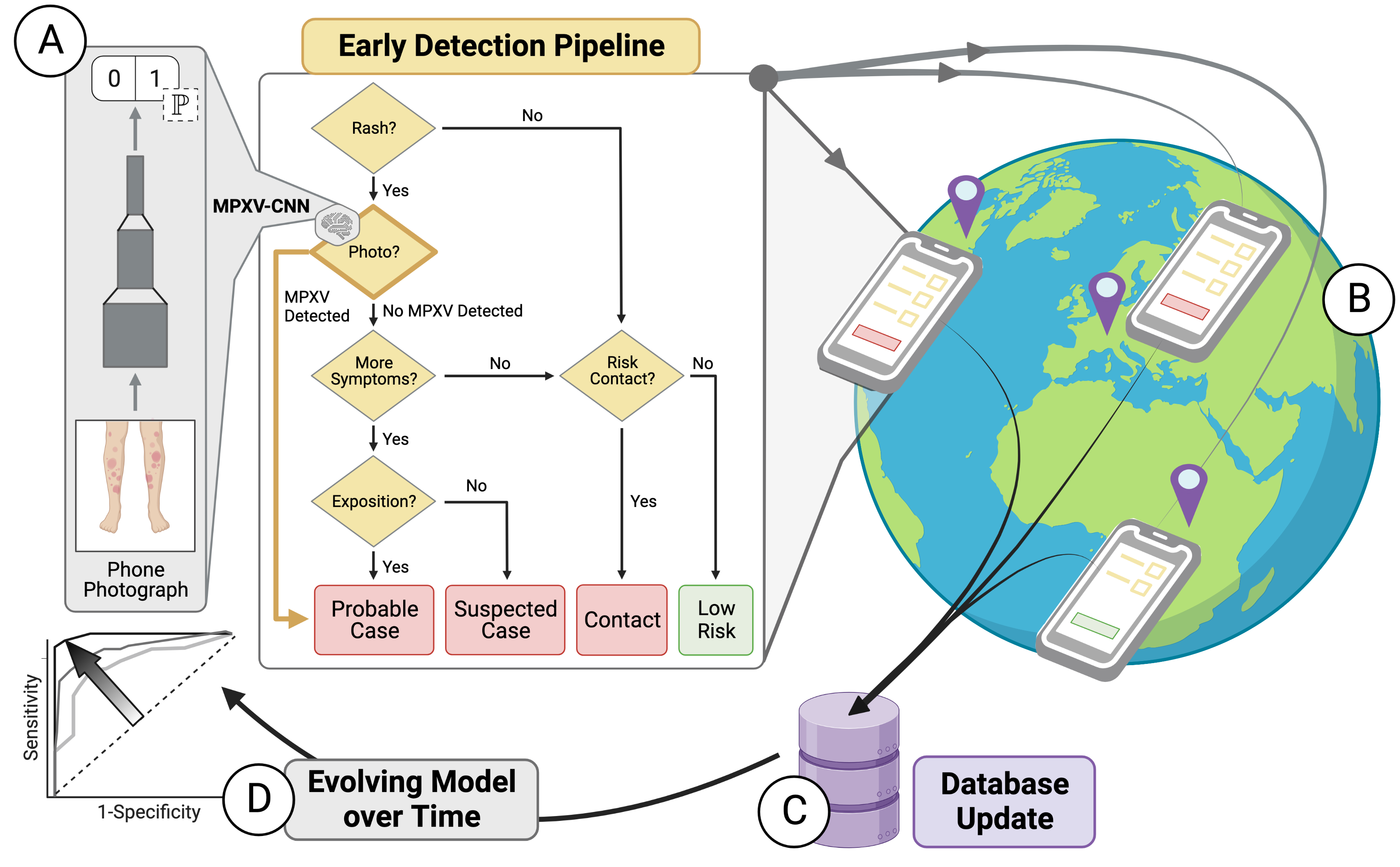
Development and Evaluation of an Image-based Deep Learning Algorithm to Classify Skin Lesions from Mpox Virus Infection
Alexander Henry Thieme, Yuanning Zheng, Gautam Machiraju, et al.
Nature Medicine (2023).
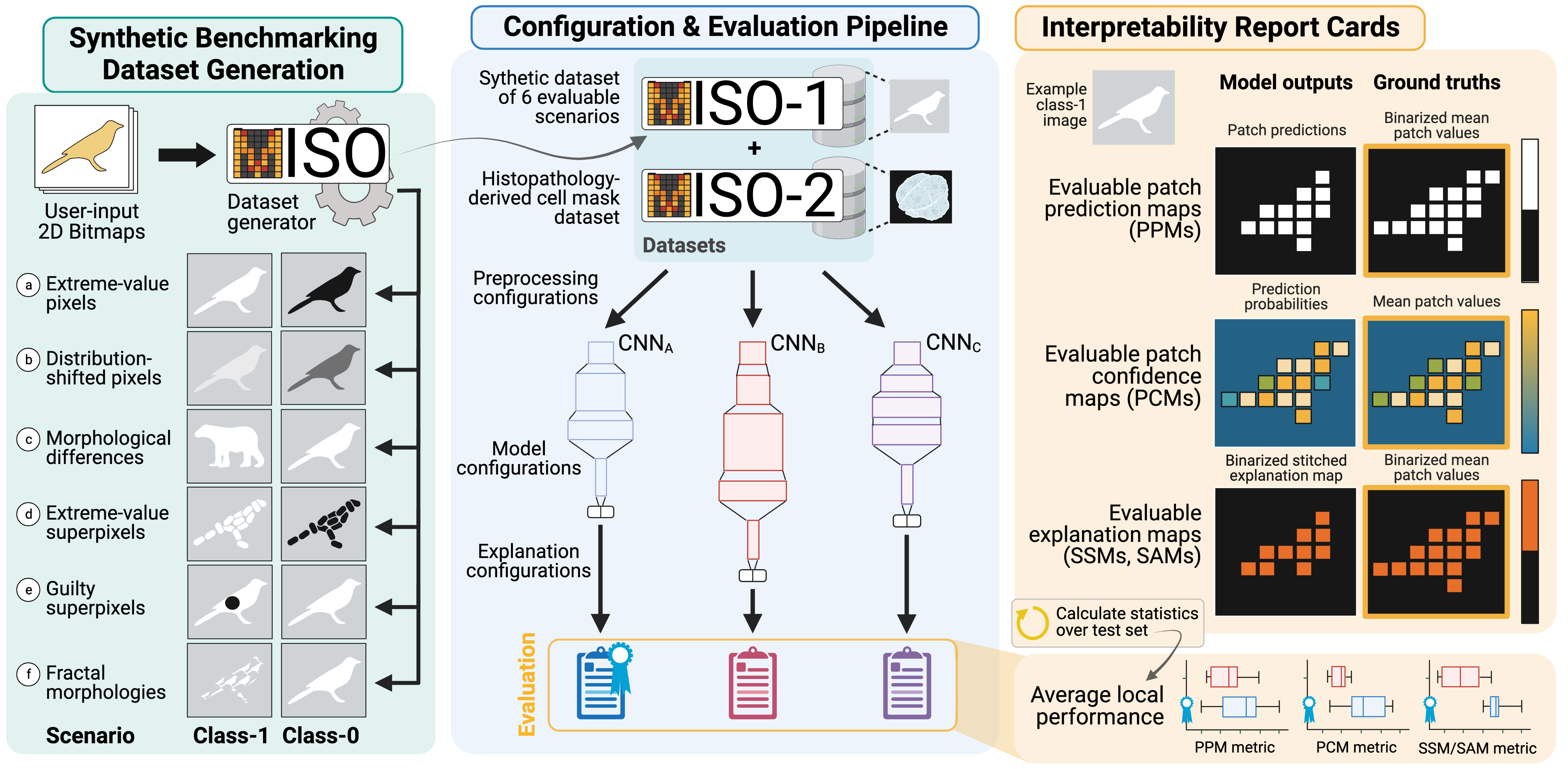
A Dataset Generation Framework for Evaluating Megapixel Image Classifiers & their Explanations
Gautam Machiraju, Sylvia Plevritis, Parag Mallick
European Conference on Computer Vision (ECCV), 2022.
Developing Machine Learning Models to Personalize Care Levels among Emergency Room Patients for Hospital Admission
Minh Nguyen, Conor Corbin, Tiffany Eulalio, Nicolai Ostberg, Gautam Machiraju, Ben Marafino, Michael Baiocchi, Christian Rose, Jonathan Chen
Journal of the American Medical Informatics Association (2021).
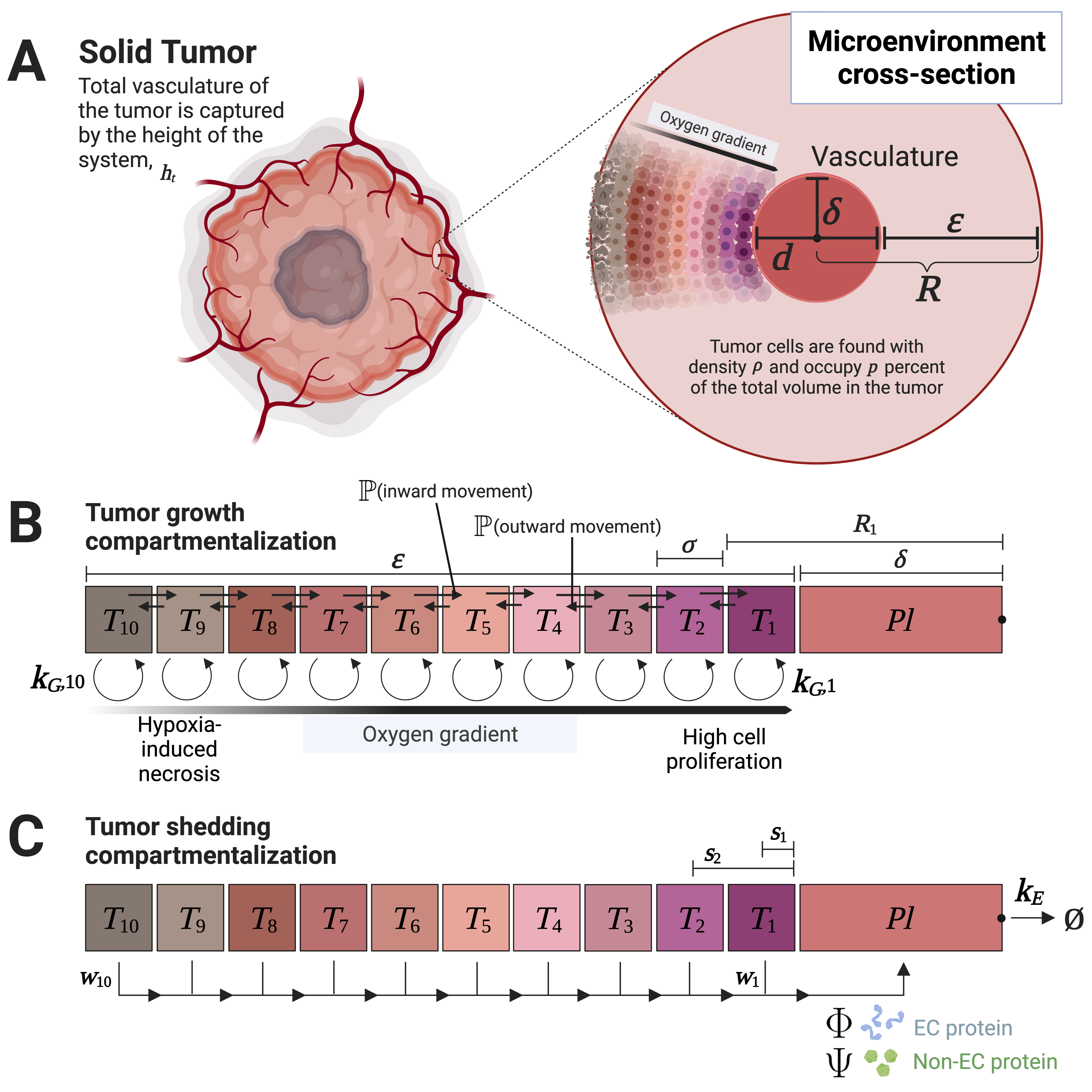
Multicompartment Modeling of Protein Shedding Kinetics During Vascularized Tumor Growth
Gautam Machiraju, Parag Mallick, Hermann Frieboes
Nature Scientific Reports (2020).
Vitæ
More details (projects, collaborators, talks, academic service, relevant coursework) can be found on my CV and Linkedin page.
-
SmarterDx 2025 - presentML Research Scientist
LLM interpretability & steering for healthcare -
Stanford AI Lab (SAIL) 2018 - 2024PhD Student
Biomedical Informatics
Data copilots for biomedical discovery -
IBM Research Summer-Fall 2023Student Researcher
Fact-checking foundation models -
Canary Center at Stanford 2016 - 2018Bioinformatics Research Assistant
Mathematical modeling of tumors,
NLP of biomedical literature,
& anomaly detection in multi-omic time-series -
Strateos Summer 2016Bioengineering SDE Intern
Robotic wet-lab automation -
University of California, Berkeley 2012 - 2016BA Student
Applied Mathematics (Math-Bio)
Minor in Bioengineering
Process
Fun: On weekends, you can find me and my partner grabbing some sun in urban-suburban East Bay. Or we're unplugging at one of California's numerous regional, state, or national parks to spend time on the water and trails. Outside of hiking and camping, my hobbies include painting, gardening, and hosting regular themed dinner and cocktail parties. I also love city trekking with friends and popping into cafés and roasteries, museums, used bookstores (hunting for vintage maths sections), and outdoor pubs with live music sessions. Reach out if you'd like to grab a coffee!
Design: One of my favorite aspects of research is thinking about aesthetic and design when communicating technical ideas. This drive to understand ideas by visually communicating them (often to myself) sparked as a dyslexic Maths undergraduate. Despite my numerous interests in Maths, I struggled to parse and conceptualize blocks of textual abstraction in modern mathematical presentation, typical of standard teaching materials. I thus relied heavily on intuition and visual proofs as mental anchors. Thanks in part to training as a CIR Scholar at Stanford's Hasso Plattner Institute of Design, I cartoon-ify almost everything I work on and will carve out time to reflect on, mock up, and refine any discussed concepts.
Acknowledgement
My graduate training and research have been graciously funded via the NIH (BD2K, NLM), the Stanford Data Science Institute (Data Science Scholarship), the International Alliance for Cancer Early Detection (Canary-ACED Graduate Fellowship), and Stanford's Institute for Human-Centered Artificial Intelligence (HAI).
This website uses the website design and template by Martin Saveski. Some stylisitc alterations were made with inspiration from Tatsunori Hashimoto.